Generalized Procrustes analysis#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.decomposition import PCA
from ktch.datasets import load_landmark_mosquito_wings
from ktch.landmark import GeneralizedProcrustesAnalysis
from ktch.plot import explained_variance_ratio_plot
Load mosquito wing landmark dataset#
from Rohlf and Slice 1990 Syst. Zool.
data_landmark_mosquito_wings = load_landmark_mosquito_wings(as_frame=True)
data_landmark_mosquito_wings.coords
| x | y | ||
|---|---|---|---|
| specimen_id | coord_id | ||
| 1 | 0 | -0.4933 | 0.0130 |
| 1 | -0.0777 | 0.0832 | |
| 2 | 0.2231 | 0.0861 | |
| 3 | 0.2641 | 0.0462 | |
| 4 | 0.2645 | 0.0261 | |
| ... | ... | ... | ... |
| 127 | 13 | -0.2028 | 0.0371 |
| 14 | 0.0490 | 0.0347 | |
| 15 | -0.0422 | 0.0204 | |
| 16 | 0.1004 | -0.0180 | |
| 17 | -0.1473 | -0.0057 |
2286 rows × 2 columns
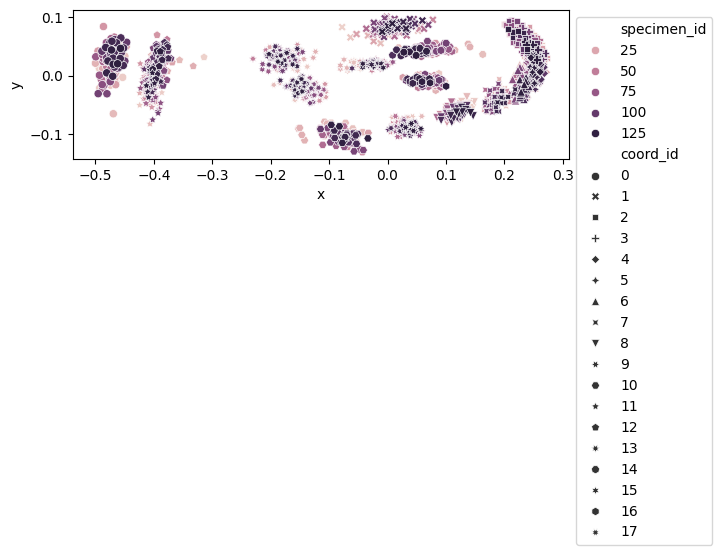
fig, ax = plt.subplots()
sns.scatterplot(
data=data_landmark_mosquito_wings.coords,
x="x",
y="y",
hue="specimen_id",
style="coord_id",
ax=ax,
)
ax.set_aspect("equal")
ax.legend(loc="upper left", bbox_to_anchor=(1, 1))
<matplotlib.legend.Legend at 0x7f1480c77850>
For applying generalized Procrustes analysis (GPA), we convert the configuration data into DataFrame of shape n_specimens x (n_landmarks*n_dim).
def configulation_plot(
configuration_2d,
x="x",
y="y",
links=[],
ax=None,
hue=None,
c="gray",
palette=None,
c_line="gray",
style=None,
s=10,
alpha=1,
):
if ax is None:
fig = plt.figure()
ax = fig.add_subplot(111)
configuration = configuration_2d.reset_index()
for link in links:
sns.lineplot(
data=configuration[configuration["coord_id"].isin(link)],
x=x,
y=y,
sort=False,
ax=ax,
hue=hue,
c=c_line,
palette=palette,
alpha=alpha,
legend=False,
)
axis = sns.scatterplot(
data=configuration,
x=x,
y=y,
ax=ax,
hue=hue,
palette=palette,
style=style,
c=c,
alpha=alpha,
s=s,
)
if axis.legend_:
sns.move_legend(ax, "upper left", bbox_to_anchor=(1, 1))
ax.set_aspect("equal")
links = [
[0, 1],
[1, 2],
[2, 3],
[3, 4],
[4, 5],
[5, 6],
[6, 7],
[7, 8],
[8, 9],
[9, 10],
[10, 11],
[1, 12],
[2, 12],
[13, 14],
[14, 3],
[14, 4],
[15, 5],
[16, 6],
[16, 7],
[17, 8],
[17, 9],
]
configulation_plot(
data_landmark_mosquito_wings.coords.loc[1], links=links, alpha=0.5, s=20
)
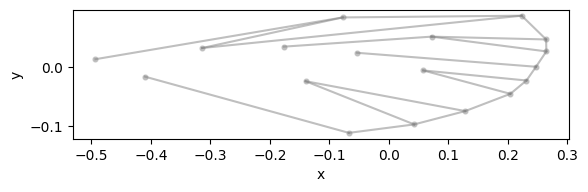
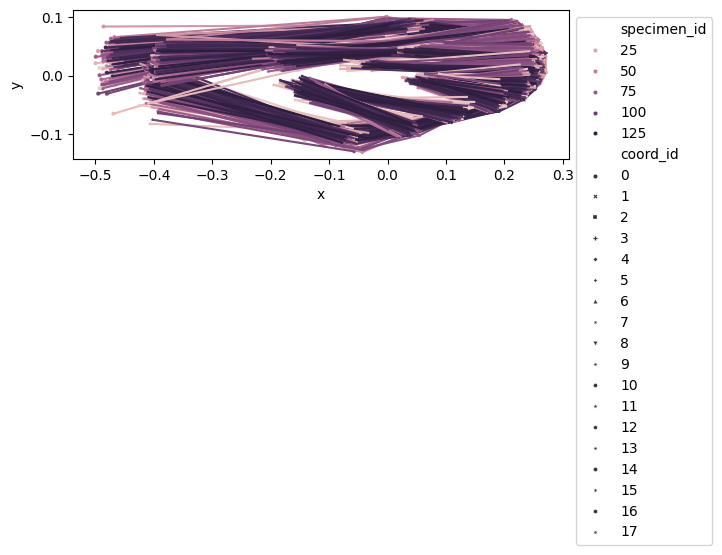
configulation_plot(
data_landmark_mosquito_wings.coords,
links=links,
hue="specimen_id",
style="coord_id",
)
configulation_plot(
data_landmark_mosquito_wings.coords.loc[0:2],
links=links,
hue="specimen_id",
style="coord_id",
palette="Set2",
s=30,
)
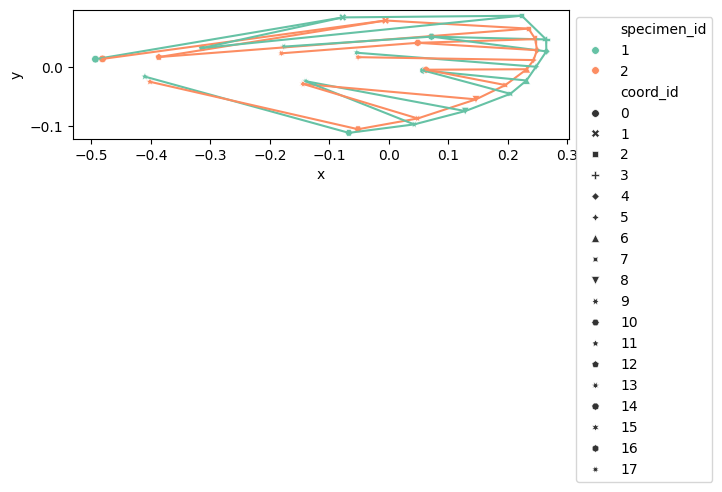
index = pd.MultiIndex.from_tuples(
[(2, i) for i in data_landmark_mosquito_wings.coords.loc[2].index],
names=["specimen_id", "coord_id"],
)
x2 = pd.DataFrame(
data_landmark_mosquito_wings.coords.loc[2].to_numpy(),
columns=["x", "y"],
index=index,
)
data_landmark_mosquito_wings.coords.loc[1:3]
| x | y | ||
|---|---|---|---|
| specimen_id | coord_id | ||
| 1 | 0 | -0.4933 | 0.0130 |
| 1 | -0.0777 | 0.0832 | |
| 2 | 0.2231 | 0.0861 | |
| 3 | 0.2641 | 0.0462 | |
| 4 | 0.2645 | 0.0261 | |
| 5 | 0.2471 | 0.0003 | |
| 6 | 0.2311 | -0.0228 | |
| 7 | 0.2040 | -0.0452 | |
| 8 | 0.1282 | -0.0742 | |
| 9 | 0.0424 | -0.0966 | |
| 10 | -0.0674 | -0.1108 | |
| 11 | -0.4102 | -0.0163 | |
| 12 | -0.3140 | 0.0318 | |
| 13 | -0.1768 | 0.0341 | |
| 14 | 0.0715 | 0.0509 | |
| 15 | -0.0540 | 0.0238 | |
| 16 | 0.0575 | -0.0059 | |
| 17 | -0.1401 | -0.0240 | |
| 2 | 0 | -0.4814 | 0.0135 |
| 1 | -0.0058 | 0.0780 | |
| 2 | 0.2345 | 0.0644 | |
| 3 | 0.2460 | 0.0467 | |
| 4 | 0.2487 | 0.0281 | |
| 5 | 0.2430 | 0.0115 | |
| 6 | 0.2316 | -0.0039 | |
| 7 | 0.1956 | -0.0305 | |
| 8 | 0.1462 | -0.0545 | |
| 9 | 0.0483 | -0.0866 | |
| 10 | -0.0520 | -0.1047 | |
| 11 | -0.4016 | -0.0250 | |
| 12 | -0.3868 | 0.0166 | |
| 13 | -0.1808 | 0.0229 | |
| 14 | 0.0484 | 0.0405 | |
| 15 | -0.0519 | 0.0164 | |
| 16 | 0.0623 | -0.0047 | |
| 17 | -0.1444 | -0.0286 | |
| 3 | 0 | -0.4622 | 0.0159 |
| 1 | 0.0089 | 0.0689 | |
| 2 | 0.2404 | 0.0545 | |
| 3 | 0.2501 | 0.0424 | |
| 4 | 0.2600 | 0.0230 | |
| 5 | 0.2541 | 0.0039 | |
| 6 | 0.2369 | -0.0105 | |
| 7 | 0.1957 | -0.0305 | |
| 8 | 0.1249 | -0.0480 | |
| 9 | 0.0146 | -0.0720 | |
| 10 | -0.0758 | -0.0865 | |
| 11 | -0.4104 | -0.0200 | |
| 12 | -0.3919 | 0.0190 | |
| 13 | -0.1724 | 0.0182 | |
| 14 | 0.0577 | 0.0344 | |
| 15 | -0.0468 | 0.0115 | |
| 16 | 0.0766 | -0.0079 | |
| 17 | -0.1602 | -0.0162 |
df_coords = (
data_landmark_mosquito_wings.coords.unstack()
.swaplevel(1, 0, axis=1)
.sort_index(axis=1)
)
df_coords.columns = [
dim + "_" + str(landmark_idx) for landmark_idx, dim in df_coords.columns
]
df_coords
| x_0 | y_0 | x_1 | y_1 | x_2 | y_2 | x_3 | y_3 | x_4 | y_4 | ... | x_13 | y_13 | x_14 | y_14 | x_15 | y_15 | x_16 | y_16 | x_17 | y_17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| specimen_id | |||||||||||||||||||||
| 1 | -0.4933 | 0.0130 | -0.0777 | 0.0832 | 0.2231 | 0.0861 | 0.2641 | 0.0462 | 0.2645 | 0.0261 | ... | -0.1768 | 0.0341 | 0.0715 | 0.0509 | -0.0540 | 0.0238 | 0.0575 | -0.0059 | -0.1401 | -0.0240 |
| 2 | -0.4814 | 0.0135 | -0.0058 | 0.0780 | 0.2345 | 0.0644 | 0.2460 | 0.0467 | 0.2487 | 0.0281 | ... | -0.1808 | 0.0229 | 0.0484 | 0.0405 | -0.0519 | 0.0164 | 0.0623 | -0.0047 | -0.1444 | -0.0286 |
| 3 | -0.4622 | 0.0159 | 0.0089 | 0.0689 | 0.2404 | 0.0545 | 0.2501 | 0.0424 | 0.2600 | 0.0230 | ... | -0.1724 | 0.0182 | 0.0577 | 0.0344 | -0.0468 | 0.0115 | 0.0766 | -0.0079 | -0.1602 | -0.0162 |
| 4 | -0.4534 | -0.0028 | -0.0318 | 0.0738 | 0.2423 | 0.0808 | 0.2627 | 0.0559 | 0.2654 | 0.0322 | ... | -0.1536 | 0.0150 | 0.0617 | 0.0436 | -0.0549 | 0.0217 | 0.0705 | -0.0031 | -0.1507 | -0.0273 |
| 5 | -0.4926 | -0.0212 | -0.0260 | 0.0708 | 0.2347 | 0.0679 | 0.2398 | 0.0584 | 0.2415 | 0.0355 | ... | -0.1374 | 0.0154 | 0.0762 | 0.0457 | -0.0313 | 0.0170 | 0.0880 | 0.0037 | -0.1318 | -0.0278 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 123 | -0.4703 | -0.0009 | 0.0011 | 0.0805 | 0.2146 | 0.0747 | 0.2348 | 0.0585 | 0.2453 | 0.0350 | ... | -0.1624 | 0.0193 | 0.0557 | 0.0467 | -0.0272 | 0.0190 | 0.0718 | -0.0052 | -0.1583 | -0.0251 |
| 124 | -0.4725 | 0.0441 | 0.0318 | 0.0808 | 0.2382 | 0.0419 | 0.2504 | 0.0237 | 0.2492 | 0.0019 | ... | -0.1724 | 0.0390 | 0.0667 | 0.0418 | -0.0108 | 0.0205 | 0.0548 | -0.0049 | -0.1546 | -0.0115 |
| 125 | -0.4697 | 0.0196 | 0.0007 | 0.0850 | 0.2228 | 0.0760 | 0.2485 | 0.0540 | 0.2548 | 0.0319 | ... | -0.1816 | 0.0213 | 0.0376 | 0.0408 | -0.0073 | 0.0137 | 0.0592 | -0.0103 | -0.1531 | -0.0300 |
| 126 | -0.4620 | 0.0204 | 0.0082 | 0.0893 | 0.2061 | 0.0797 | 0.2374 | 0.0529 | 0.2457 | 0.0306 | ... | -0.1876 | 0.0241 | 0.0279 | 0.0403 | -0.0425 | 0.0145 | 0.0729 | -0.0123 | -0.1460 | -0.0269 |
| 127 | -0.4570 | 0.0468 | -0.0090 | 0.0753 | 0.2381 | 0.0463 | 0.2556 | 0.0275 | 0.2614 | 0.0064 | ... | -0.2028 | 0.0371 | 0.0490 | 0.0347 | -0.0422 | 0.0204 | 0.1004 | -0.0180 | -0.1473 | -0.0057 |
127 rows × 36 columns
GPA#
gpa = GeneralizedProcrustesAnalysis().set_output(transform="pandas")
df_shapes = gpa.fit_transform(df_coords)
df_shapes
| x_0 | y_0 | x_1 | y_1 | x_2 | y_2 | x_3 | y_3 | x_4 | y_4 | ... | x_13 | y_13 | x_14 | y_14 | x_15 | y_15 | x_16 | y_16 | x_17 | y_17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| specimen_id | |||||||||||||||||||||
| 1 | -0.488913 | 0.021159 | -0.075651 | 0.083813 | 0.222655 | 0.081656 | 0.262641 | 0.041408 | 0.262701 | 0.021471 | ... | -0.174735 | 0.036786 | 0.071747 | 0.049290 | -0.053145 | 0.024519 | 0.056916 | -0.006796 | -0.139317 | -0.021437 |
| 2 | -0.479850 | 0.030667 | -0.002995 | 0.078029 | 0.236289 | 0.055873 | 0.247131 | 0.037801 | 0.249161 | 0.019145 | ... | -0.179578 | 0.029304 | 0.049746 | 0.038675 | -0.051194 | 0.018213 | 0.062000 | -0.006922 | -0.145098 | -0.023383 |
| 3 | -0.461238 | 0.021194 | 0.009668 | 0.068674 | 0.240606 | 0.051631 | 0.250150 | 0.039440 | 0.259809 | 0.019959 | ... | -0.171907 | 0.020150 | 0.057987 | 0.033671 | -0.046599 | 0.012014 | 0.076367 | -0.008775 | -0.160124 | -0.014331 |
| 4 | -0.451582 | 0.021733 | -0.027671 | 0.075199 | 0.245617 | 0.067345 | 0.264582 | 0.041450 | 0.265988 | 0.017707 | ... | -0.152122 | 0.023242 | 0.063790 | 0.040074 | -0.053488 | 0.024575 | 0.070026 | -0.006899 | -0.151522 | -0.019031 |
| 5 | -0.491092 | 0.019428 | -0.020015 | 0.072475 | 0.238724 | 0.048169 | 0.243009 | 0.038313 | 0.242816 | 0.015425 | ... | -0.135231 | 0.026596 | 0.079447 | 0.039142 | -0.029701 | 0.019466 | 0.087717 | -0.003550 | -0.133219 | -0.016779 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 123 | -0.468838 | 0.028646 | 0.006143 | 0.080164 | 0.218584 | 0.060969 | 0.237700 | 0.043553 | 0.246690 | 0.019469 | ... | -0.160671 | 0.029436 | 0.058442 | 0.043044 | -0.025929 | 0.020642 | 0.071228 | -0.009699 | -0.159374 | -0.015077 |
| 124 | -0.473804 | 0.016670 | 0.027045 | 0.082425 | 0.235150 | 0.055559 | 0.248370 | 0.038112 | 0.248433 | 0.016300 | ... | -0.174200 | 0.028931 | 0.064108 | 0.045546 | -0.011957 | 0.019822 | 0.054939 | -0.001719 | -0.153528 | -0.020407 |
| 125 | -0.468449 | 0.032047 | 0.002958 | 0.084844 | 0.224473 | 0.069955 | 0.249549 | 0.047306 | 0.255251 | 0.025073 | ... | -0.180752 | 0.026088 | 0.038626 | 0.039732 | -0.006925 | 0.013867 | 0.058834 | -0.011863 | -0.153659 | -0.025890 |
| 126 | -0.459471 | 0.036434 | 0.011277 | 0.088662 | 0.208061 | 0.072197 | 0.238303 | 0.044411 | 0.245792 | 0.021909 | ... | -0.186025 | 0.030549 | 0.029190 | 0.039168 | -0.041832 | 0.015925 | 0.072178 | -0.014794 | -0.146368 | -0.021701 |
| 127 | -0.457745 | 0.025260 | -0.012492 | 0.074647 | 0.235186 | 0.057309 | 0.253512 | 0.039387 | 0.260281 | 0.018625 | ... | -0.203892 | 0.027493 | 0.047226 | 0.036891 | -0.043017 | 0.018365 | 0.100931 | -0.013236 | -0.146563 | -0.012573 |
127 rows × 36 columns
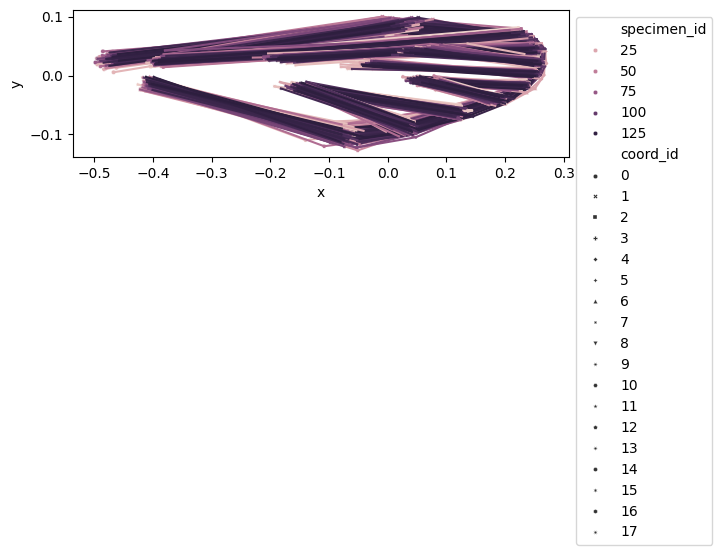
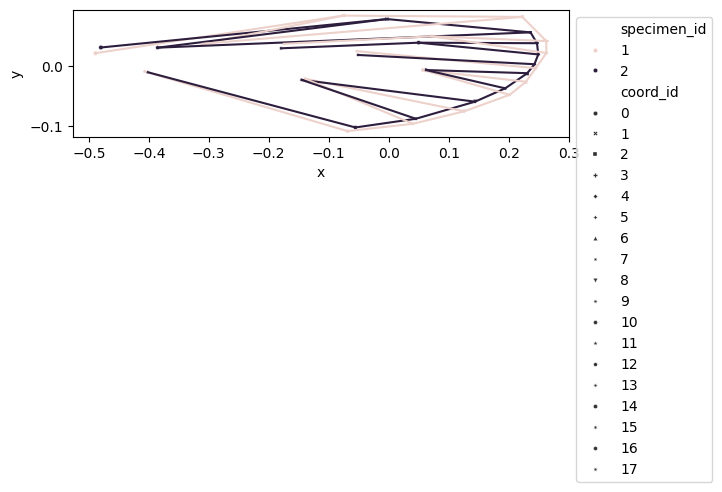
We create a DataFrame, called df_shapes_vis, of shape (n_specimens*n_landmarks) x n_dim to visualize the aligned shapes.
df_shapes_vis = df_shapes.copy()
df_shapes_vis.columns = pd.MultiIndex.from_tuples(
[
[int(landmark_idx), dim]
for dim, landmark_idx in [idx.split("_") for idx in df_shapes_vis.columns]
],
names=["coord_id", "dim"],
)
df_shapes_vis.sort_index(axis=1, inplace=True)
df_shapes_vis = df_shapes_vis.swaplevel(0, 1, axis=1).stack(level=1, future_stack=True)
df_shapes_vis
| dim | x | y | |
|---|---|---|---|
| specimen_id | coord_id | ||
| 1 | 0 | -0.488913 | 0.021159 |
| 1 | -0.075651 | 0.083813 | |
| 2 | 0.222655 | 0.081656 | |
| 3 | 0.262641 | 0.041408 | |
| 4 | 0.262701 | 0.021471 | |
| ... | ... | ... | ... |
| 127 | 13 | -0.203892 | 0.027493 |
| 14 | 0.047226 | 0.036891 | |
| 15 | -0.043017 | 0.018365 | |
| 16 | 0.100931 | -0.013236 | |
| 17 | -0.146563 | -0.012573 |
2286 rows × 2 columns
fig, ax = plt.subplots()
sns.scatterplot(
data=df_shapes_vis, x="x", y="y", hue="specimen_id", style="coord_id", ax=ax
)
ax.set_aspect("equal")
ax.legend(loc="upper left", bbox_to_anchor=(1, 1))
<matplotlib.legend.Legend at 0x7f1475408f10>
configulation_plot(df_shapes_vis, links=links, hue="specimen_id", style="coord_id")
configulation_plot(
df_shapes_vis.loc[0:2], links=links, hue="specimen_id", style="coord_id"
)
PCA#
pca = PCA(n_components=10).set_output(transform="pandas")
df_pca = pca.fit_transform(df_shapes)
df_pca = df_pca.join(data_landmark_mosquito_wings.meta)
df_pca
| pca0 | pca1 | pca2 | pca3 | pca4 | pca5 | pca6 | pca7 | pca8 | pca9 | genus | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| specimen_id | |||||||||||
| 1 | -0.003856 | -0.093210 | 0.029908 | 0.049589 | -0.007598 | 0.042657 | -0.005370 | 0.010359 | -0.011595 | 0.034991 | AN |
| 2 | -0.029857 | -0.023434 | 0.006114 | -0.017268 | -0.014229 | 0.014271 | 0.013344 | 0.021849 | -0.011211 | 0.005069 | AN |
| 3 | -0.012572 | -0.019728 | -0.034610 | -0.025665 | -0.005116 | -0.017905 | 0.008511 | -0.004312 | 0.001597 | 0.009884 | AN |
| 4 | -0.001652 | -0.049890 | -0.034137 | -0.001013 | 0.019706 | -0.010102 | -0.001692 | -0.010425 | 0.021595 | -0.006472 | AN |
| 5 | 0.026869 | -0.032989 | 0.010228 | -0.051852 | 0.016741 | 0.023749 | 0.015917 | -0.000273 | -0.013100 | 0.012755 | AN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 123 | -0.007864 | -0.009853 | -0.006369 | -0.026082 | -0.025424 | 0.013786 | -0.009967 | -0.018669 | 0.000513 | -0.005515 | CX |
| 124 | -0.005360 | 0.014451 | -0.014347 | -0.013199 | 0.000102 | 0.001379 | -0.002919 | -0.009755 | -0.020409 | -0.017790 | CX |
| 125 | -0.030056 | 0.000885 | 0.013455 | 0.000542 | -0.008940 | 0.008503 | -0.021390 | -0.014563 | -0.003408 | -0.010083 | CX |
| 126 | -0.048757 | -0.000199 | 0.024147 | -0.029841 | -0.040190 | 0.013335 | 0.000598 | 0.016086 | 0.018985 | 0.004560 | DE |
| 127 | -0.017155 | -0.030018 | -0.001935 | -0.014438 | -0.014540 | -0.035565 | -0.011888 | 0.017324 | -0.009739 | 0.012342 | DE |
127 rows × 11 columns
fig, ax = plt.subplots()
sns.scatterplot(data=df_pca, x="pca0", y="pca1", hue="genus", palette="Paired", ax=ax)
ax.legend(loc="upper left", bbox_to_anchor=(1, 1))
<matplotlib.legend.Legend at 0x7f14725d4590>
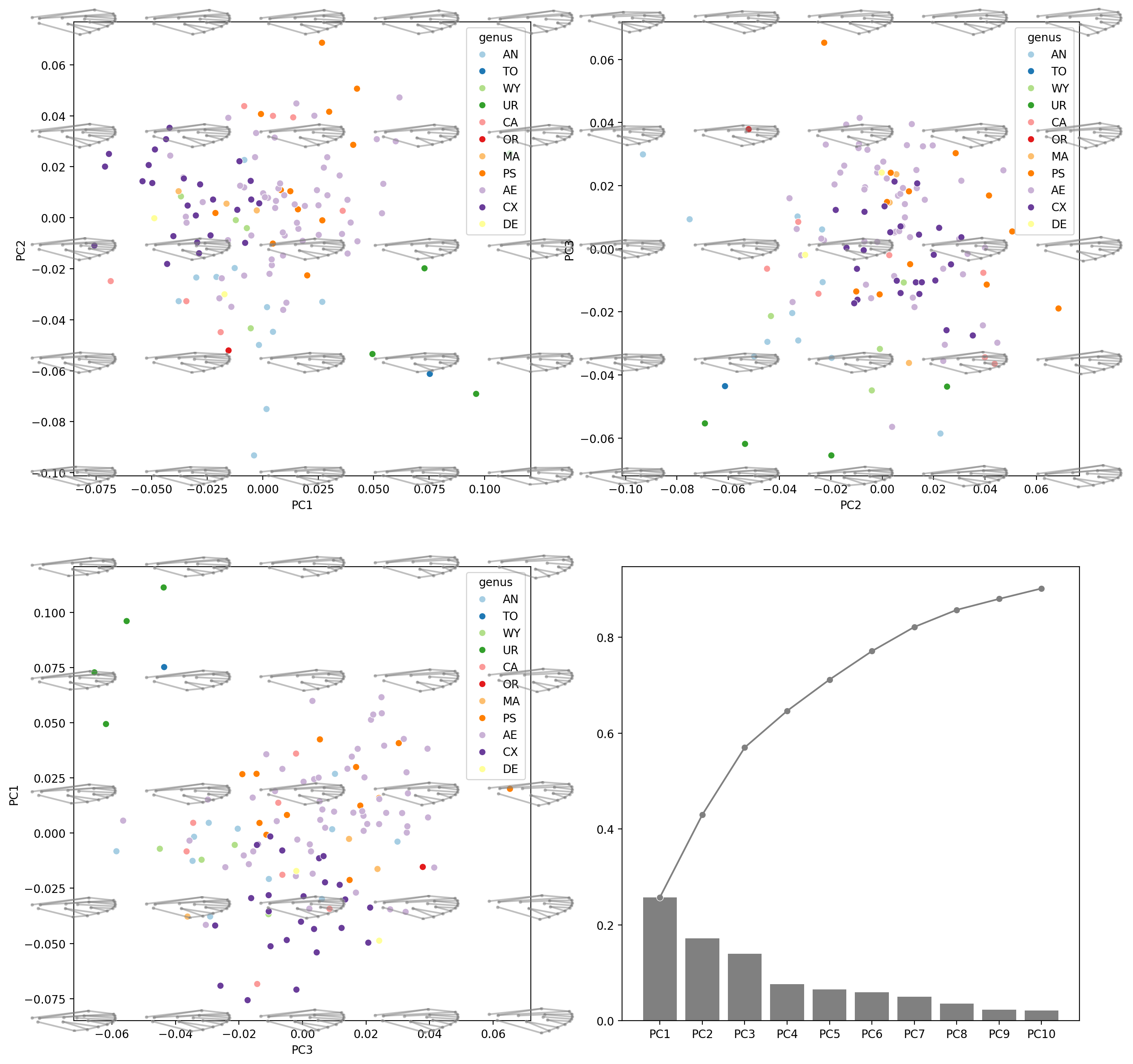
Morphospace#
def get_pc_scores_for_morphospace(ax, num=5):
xrange = np.linspace(ax.get_xlim()[0], ax.get_xlim()[1], num)
yrange = np.linspace(ax.get_ylim()[0], ax.get_ylim()[1], num)
return xrange, yrange
def plot_recon_morphs(
pca,
fig,
ax,
n_PCs_xy=[1, 2],
morph_num=3,
morph_alpha=1.0,
morph_scale=1.0,
links=[],
):
pc_scores_h, pc_scores_v = get_pc_scores_for_morphospace(ax, morph_num)
print("PC_h: ", pc_scores_h, ", PC_v: ", pc_scores_v)
for pc_score_h in pc_scores_h:
for pc_score_v in pc_scores_v:
pc_score = np.zeros(pca.n_components_)
n_PC_h, n_PC_v = n_PCs_xy
pc_score[n_PC_h - 1] = pc_score_h
pc_score[n_PC_v - 1] = pc_score_v
arr_shapes = pca.inverse_transform([pc_score])
arr_shapes = arr_shapes.reshape(-1, 2)
df_shapes = pd.DataFrame(arr_shapes, columns=["x", "y"])
df_shapes["coord_id"] = [i for i in range(len(df_shapes))]
df_shapes = df_shapes.set_index("coord_id")
ax_width = ax.get_window_extent().width
fig_width = fig.get_window_extent().width
fig_height = fig.get_window_extent().height
morph_size = morph_scale * ax_width / (fig_width * morph_num)
loc = ax.transData.transform((pc_score_h, pc_score_v))
if arr_shapes.shape[1] == 3:
axins = fig.add_axes(
[
loc[0] / fig_width - morph_size / 2,
loc[1] / fig_height - morph_size / 2,
morph_size,
morph_size,
],
anchor="C",
projection="3d",
)
axins.patch.set_alpha(0.3)
configulation_plot(df_shapes, links=links, ax=axins, alpha=morph_alpha)
else:
axins = fig.add_axes(
[
loc[0] / fig_width - morph_size / 2,
loc[1] / fig_height - morph_size / 2,
morph_size,
morph_size,
],
anchor="C",
)
configulation_plot(df_shapes, links=links, ax=axins, alpha=morph_alpha)
axins.axis("off")
morph_num = 5
morph_scale = 1.0
morph_alpha = 0.5
fig = plt.figure(figsize=(16, 16), dpi=200)
hue_order = df_pca["genus"].unique()
#########
# PC1
#########
ax = fig.add_subplot(2, 2, 1)
sns.scatterplot(
data=df_pca,
x="pca0",
y="pca1",
hue="genus",
hue_order=hue_order,
palette="Paired",
ax=ax,
legend=True,
)
plot_recon_morphs(
pca,
morph_num=5,
morph_scale=morph_scale,
morph_alpha=0.5,
fig=fig,
ax=ax,
links=links,
)
ax.patch.set_alpha(0)
ax.set(xlabel="PC1", ylabel="PC2")
print("PC1-PC2 done")
#########
# PC2
#########
ax = fig.add_subplot(2, 2, 2)
sns.scatterplot(
data=df_pca,
x="pca1",
y="pca2",
hue="genus",
hue_order=hue_order,
palette="Paired",
ax=ax,
legend=True,
)
plot_recon_morphs(
pca,
morph_num=5,
morph_scale=morph_scale,
morph_alpha=0.5,
fig=fig,
ax=ax,
links=links,
n_PCs_xy=[2, 3],
)
ax.patch.set_alpha(0)
ax.set(xlabel="PC2", ylabel="PC3")
print("PC2-PC3 done")
#########
# PC3
#########
ax = fig.add_subplot(2, 2, 3)
sns.scatterplot(
data=df_pca,
x="pca2",
y="pca0",
hue="genus",
hue_order=hue_order,
palette="Paired",
ax=ax,
legend=True,
)
plot_recon_morphs(
pca,
morph_num=5,
morph_scale=morph_scale,
morph_alpha=0.5,
fig=fig,
ax=ax,
links=links,
n_PCs_xy=[3, 1],
)
ax.patch.set_alpha(0)
ax.set(xlabel="PC3", ylabel="PC1")
print("PC3-PC1 done")
#########
# CCR
#########
ax = fig.add_subplot(2, 2, 4)
explained_variance_ratio_plot(pca, ax=ax, verbose=True)
PC_h: [-0.08507944 -0.03363238 0.01781469 0.06926176 0.12070883] , PC_v: [-0.10130283 -0.05679242 -0.01228202 0.03222838 0.07673878]
PC1-PC2 done
PC_h: [-0.10130283 -0.05679242 -0.01228202 0.03222838 0.07673878] , PC_v: [-7.20613317e-02 -3.60802028e-02 -9.90738801e-05 3.58820550e-02
7.18631840e-02]
PC2-PC3 done
PC_h: [-7.20613317e-02 -3.60802028e-02 -9.90738801e-05 3.58820550e-02
7.18631840e-02] , PC_v: [-0.08507944 -0.03363238 0.01781469 0.06926176 0.12070883]
PC3-PC1 done
Explained variance ratio:
['PC1 0.25758090980106463', 'PC2 0.17223024539391402', 'PC3 0.1402677835736617', 'PC4 0.0763462878404697', 'PC5 0.06525408598811429', 'PC6 0.05936398220741054', 'PC7 0.05041804355293129', 'PC8 0.03566678305170681', 'PC9 0.022911799413012413', 'PC10 0.02174073028592705']
Cumsum of Explained variance ratio:
['PC1 0.25758090980106463', 'PC2 0.42981115519497864', 'PC3 0.5700789387686404', 'PC4 0.6464252266091101', 'PC5 0.7116793125972244', 'PC6 0.771043294804635', 'PC7 0.8214613383575663', 'PC8 0.8571281214092731', 'PC9 0.8800399208222855', 'PC10 0.9017806511082126']
<Axes: >